Reading on Pandas DataFrame
Motivation for the blog
So I got a little overwhelmed with a lot of ways we can CRUD on DataFrame and which to use when. I am writing this blog to gather my understanding and make it concrete.
Context for developers
DataFrame is a data structure available in the pandas library in python. You will see how do we use it now.
You can start thinking about the DataFrame as a tabular data structure. Much like in spreadsheets where each cell can be identified with the help of a (row,column). If you aware of a python list and coming to the level of list of lists which makes it a tabular data structure, let’s create that first so that we can appreciate what DataFrames offer.
food = [['apple', 'banana', 'pineapple'],
['potato', 'cabbage', 'cauliflower', 'eggplant'],
['wheat', 'oats', 'rice'],
['kidney beans', 'chickpeas', 'soybeans']]We know in our minds that the first row is for “fruits”, then “vegetables”, then “grains”, then “legumes”. But this information is absent in the above list of lists. Accessing a particular food will require you to already being aware about which food is at which index. for example, If we need “fruits”, you should know that food[0] will give you all “fruits”; food[2][0] gives you “wheat”.
The next problem seems to be solved by using a dictionary object where we can define the same data as
food = {
"fruits": ['apple', 'banana', 'pineapple'],
"vegetables": ['potato', 'cabbage', 'cauliflower', 'eggplant'],
"grains": ['wheat', 'oats', 'rice'],
"legumes": ['kidney beans', 'chickpeas', 'soybeans']
}
food["fruits"] // ['apple', 'banana', 'pineapple']With the dictionary data structure we can now access fruits by the lable name. If you notice, we can have different number of fruits, let’s say 10 fruits, and 12 vegetables. Both lists & dictionary data structure allow it.
- Lists did not allow labelling of rows and columns.
- Dictionary doesn’t allow reaching an element by number indexes.
- List of lists & Dictionary doesn’t guarantee to be table-like since it allows different length of child lists
---------------------------------------------------------------------------
KeyError Traceback (most recent call last)
Cell In[10], line 1
----> 1 food[0]
KeyError: 0Why do we care about tabular structure so much?
Think about a domain or an industry, let’s take the case of food itself first so that we can connect. The example data above mentions catagorical data. The number of categories are limited, there can not be millions of categories of food. If I try to make a tabular data in a spreadsheet, the columns can be fruits, vegetables, grains, legumes. These columns are independent of each other. Also, picking a row in the table doesn’t make much sense. Let’s visualise it a little.
| fruits | vegetables | grains | legumes | |
| 1 | apple | potato | wheat | kidney beans |
| 2 | banana | cabbage | oats | chickpeas |
| 3 | pineapple | cauliflower | rice | soybeans |
| 4 | eggplant |
What kind of data would make sense? Let’s take this data
| fruits | vegetables | grains | legumes | |
| 1 | apple | potato | wheat | kidney beans |
| 2 | apple | cabbage | rice | soybeans |
| 3 | banana | cauliflower | rice | soybeans |
| 4 | apple | potato | oats | chickpeas |
| 5 | pineapple | eggplant | rice | soybeans |
| 6 | banana | potato | wheat | kidney beans |
| 7 | banana | eggplant | oats | chickpeas |
| 8 | orange | french beans | NA | chickpeas |
| 9 | kiwi | NA | ragi | NA |
Assume, each row is the meal intake of a person. This is not categorical data. Each row means something. Also assume, we collect this data for an entire neighbourhood, the number of rows can keep increasing with time. Add a few columns like id, name and scale the data to an entire country, the number of rows can be in millions.
This kind of tabular data makes sense. Each row is an event. Each column can be thought of as important details of the event (a.k.a dimensions). We can add more important-details-of-the-event by adding more columns. Let’s say we add a column called drink, or protein (chicken, fish, egg, etc.)
list of lists & dictonary data structures can hold this tabular data too. We will be able to do all sort of processing on it too. But we have to write a lot of efficient code, many times to CRUD, manipulate the data. This problem is solved by python libraries like numpy and pandas. These libraries provide data structures like Series and DataFrame to easily read/manipulate the data, combine the data, do sql like joins, choose smaller frames of the data (and manipulate it too). etc.
DataFrame
| fruits | vegetables | grains | legumes | |
| 0 | apple | potato | wheat | kidney beans |
| 1 | apple | cabbage | rice | soybeans |
| 2 | banana | cauliflower | rice | soybeans |
| 3 | apple | potato | oats | chickpeas |
| 4 | pineapple | eggplant | rice | soybeans |
| 5 | banana | potato | wheat | kidney beans |
| 6 | banana | eggplant | oats | chickpeas |
| 7 | orange | french beans | NA | chickpeas |
| 8 | kiwi | NA | ragi | NA |
It’s a tabular data structure. The first horizontal block (here fruits vegetables grains legumes) are the columns. The first vertical block (here 0 1 2 3 4 5 6 7 8) are the index.
We will talk about creating a DataFrame a little later, for now let’s talk about the more interesting part of reading the data.
What do you want to read? This is the main topic of this blog.
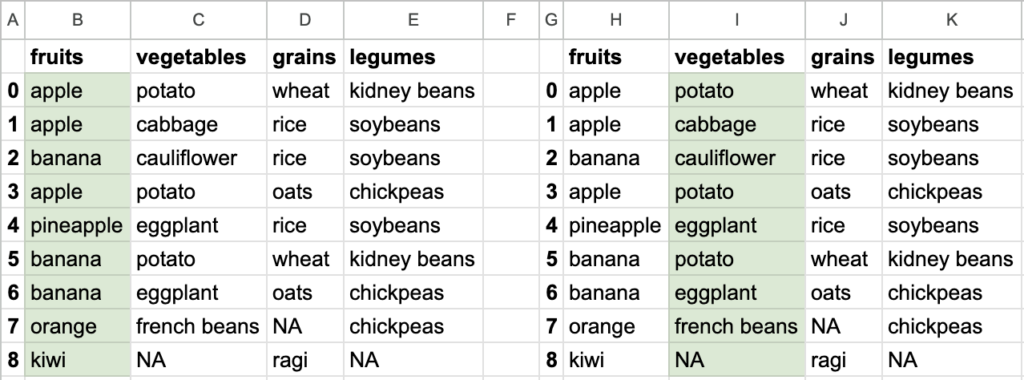
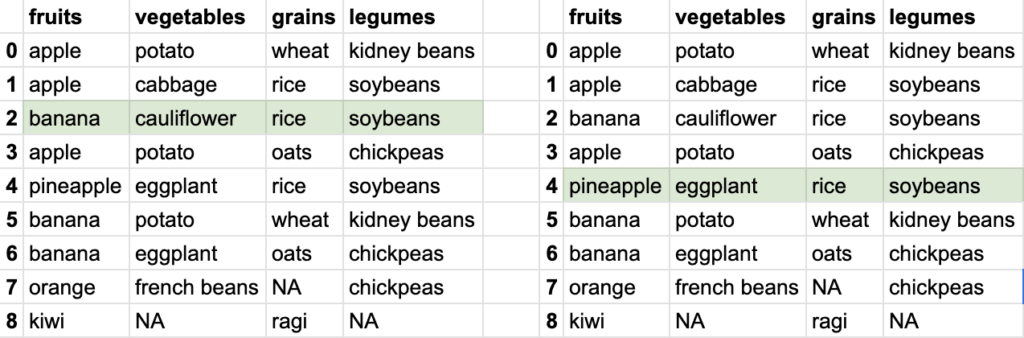
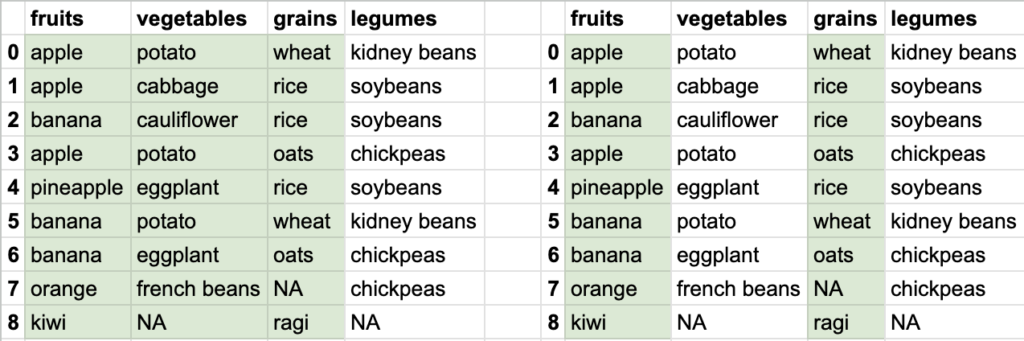
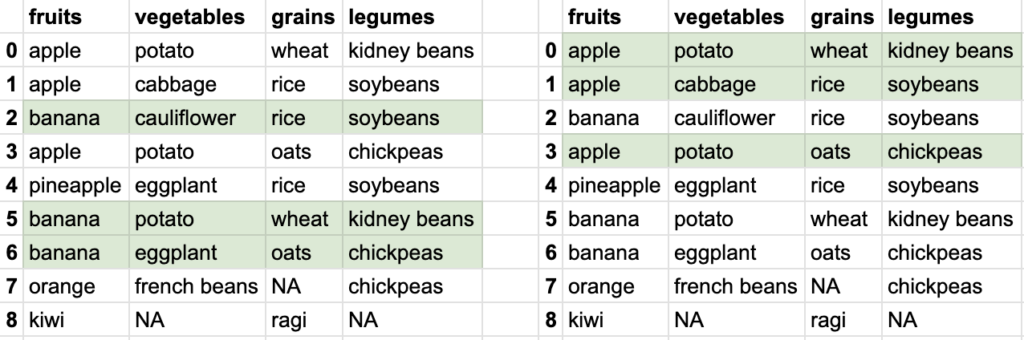
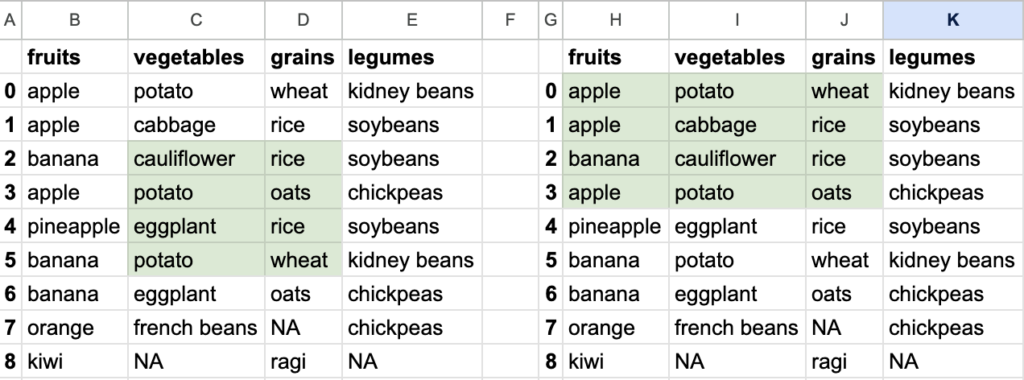
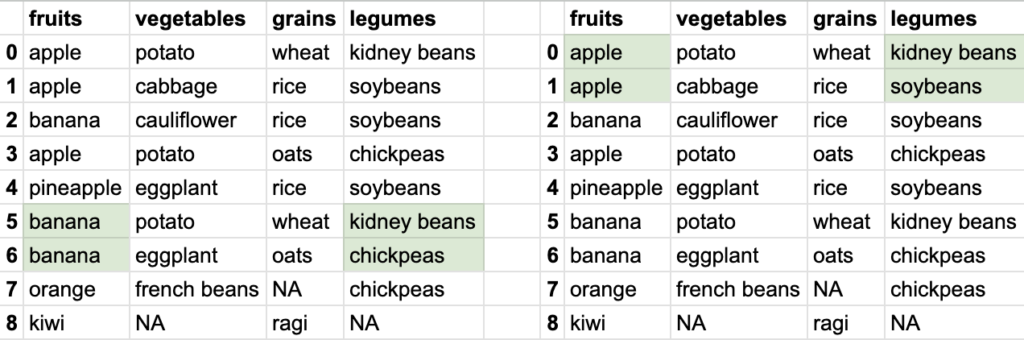
So you see how different data can be extracted from the bigger tabular data. Tabular data stored in DataFrame allows ways to extract smaller datasets. Of course, DataFrame is capable of doing a lot of other things, but let’s focus on reading the data first.
What methods are available to read smaller datasets?
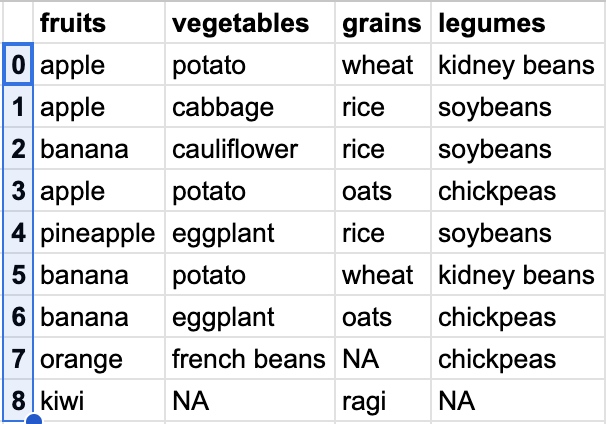
Let’s define our DataFrame. Remember we will talk about defining the DataFrame later. This is just one way to define it.
meal = [['apple', 'potato', 'wheat', 'kidney beans'],
['apple', 'cabbage', 'rice', 'soybeans'],
['banana', 'cauliflower', 'rice', 'soybeans'],
['apple', 'potato', 'oats', 'chickpeas'],
['pineapple', 'eggplant', 'rice', 'soybeans'],
['banana', 'potato', 'wheat', 'kidney beans'],
['banana', 'eggplant', 'oats', 'chickpeas'],
['orange', 'french beans', None, 'chickpeas'],
['kiwi', None, 'ragi', None]]
df = pd.DataFrame(meal, columns=['fruits', 'vegetables', 'grains', 'legumes'])This is how it looks
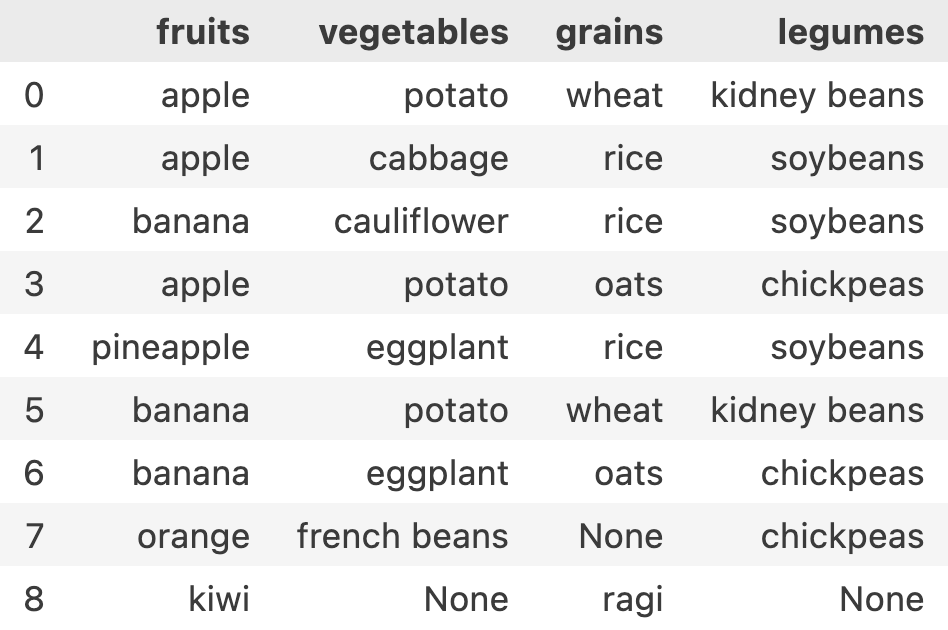
Choosing specific column(s), all rows
fruits = df['fruits']
print(type(fruits)) // <class 'pandas.core.series.Series'>
A Series data structure allows accessing specific element by regular indexing fruits[0] // 'apple'
Accessing a single column on a
DataFramevia the syntaxdf['column name']will result in aSeries
fruits = df[['fruits']]
print(type(fruits)) // <class 'pandas.core.frame.DataFrame'>
fruits_and_grains = df[['fruits', 'grains']]
print(type(fruits_and_grains)) // <class 'pandas.core.frame.DataFrame'>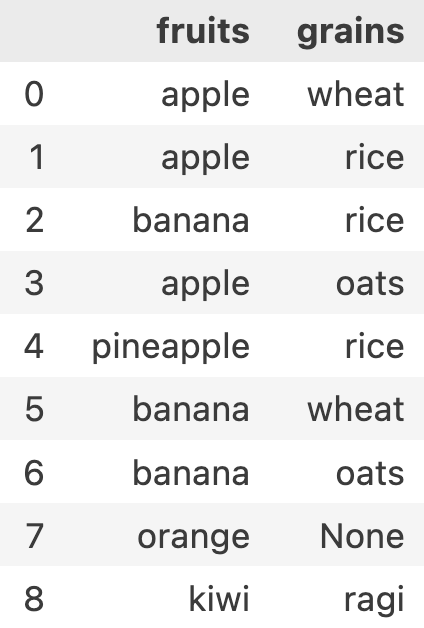
Accessing one or more column on a DataFrame via syntax
df[['column name', 'column name']]will result in aDataFrame
The important point to note here is that you are not just choosing specific columns. You are also choosing all the rows of those specific columns. If you need all the rows, go for this syntax df['column name'] or df[['column name', column name']]. Let’s call this column indexing.
Choosing a specific cell by row-number, column-number
This looks like another simple case. DataFrame doesn’t allow syntax like df[row-number, column-number], rather it allows this via a method called iloc. The syntax will look like df.iloc[row-number, column-number]. The column fruits in the DataFrame df is column-number 0, and so on.
If I want to find what legume(index 3, since 0 based indexing) was eaten in the 5th meal(index 4, since 0 based indexing), i’ll go for syntax like df.iloc[4, 3], which results in soybeans. check in the image below.

Choosing a smaller frame in the DataFrame
The interesting thing about iloc is that it allows another syntax i.e. df.iloc['row-range', 'column-range']. If you remember slicing in python lists, the row-range, column-range will follow the same format.
- “
:” represents all - “
1:” represents that leave the one at 0th index and take till last - “
:5” represents that start from the 0th index and take till 4th index - and so on
If I want to get fruits, vegetables, grains, legumes for 3rd till 7th index, my row-range will be 3:8 and my column-range will be : since I want to fetch all columns. entire syntax will be df.iloc[3:8, :]
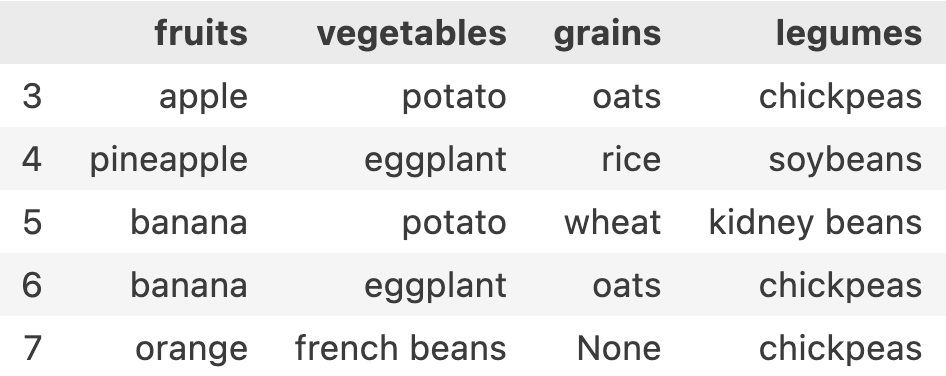
if I want to get vegetables and grains for 3rd till 7th index, my row-range is still 3:8 and my new column-range will be 1:3. entire syntax will be df.iloc[3:8, 1:3]
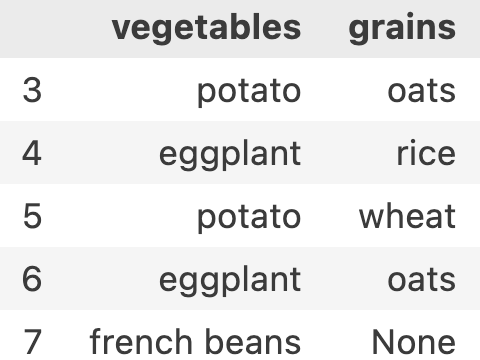
If you remember in python list slicing, there was also an option to provide step. let’s try giving a step. If I want to get all even number meals, my row-range will look like ::2. Now df.iloc[::2, 1:3] results in
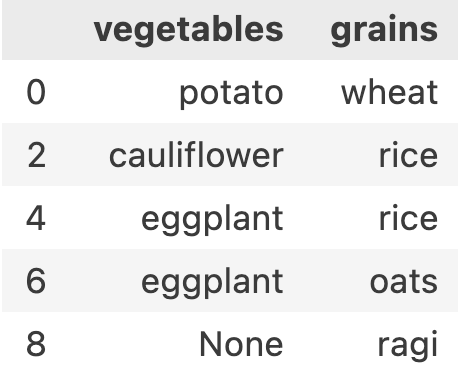
iloc will be useful when we have enough information about the digit of the row or column to be chosen.
Choosing a specific cell by row-name, column-name
When I use the word row-name i mean the the first vertical block of the DataFrame i.e. 0 1 2 3 4 5 6 7 8. In the terminology of DataFrame these are not really integer digits, they are “names”. Specifically called index. (for the curious, I can have indexes like a b c d e or india china srilanka. it’s also possible to have multiple indexes, but that is out of scope of this discussion)
When I use the word column-name. It’s simple, they are the column names, i.e. fruits vegetables grains legumes.
If I want to find what legume was consumed in meal name 5, we use the syntax df.loc[5, 'legume'] it results in kidney beans
Choosing specific cells by row-name-array, column-name-array
loc allows another syntax where I can pass array of row-names and array of column-names. i.e. df.loc[[3, 5, 6], ['vegetables', 'legumes']]
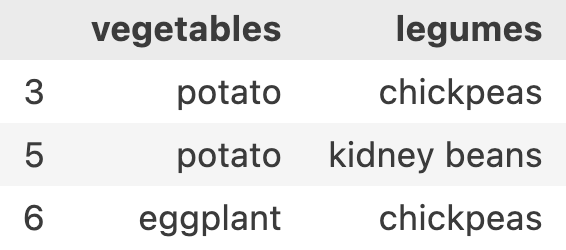
This is where it becomes a little bit more interesting. Sometimes, we don’t know the exact row-names and column-names to choose because there can be millions of rows and hundreds of columns.
Assume there are millions of rows and we can filter them based on a condition. Similarly let’s say there are hundreds of columns and we can choose specific columns by filtering on their names, let’s say we choose all the columns that start with solid_ for solid food and fluid_ for fluid foods.
In our example data, we have 9 meals. Let’s say I am able to somehow create an array for those 9 meals like so [False, False, False, True, False, True, True, False, False]. This array indicates that row-name 3, 5 & 6 are to be chosen since only they are true.
Let’s see what this syntax does df.loc[[False, False, False, True, False, True, True, False, False], ['vegetables', legumes']]
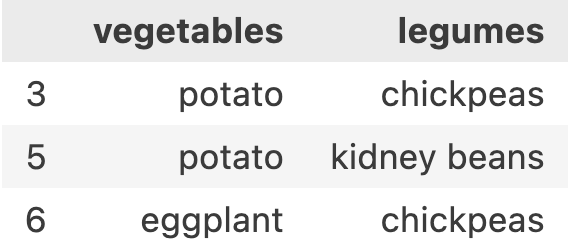
It gave me the same result as df.loc[[3,5,6], ['vegetables', 'legumes']]. We will talk about it a little later how passing multiple trues and falses are more interesting to us.
Similarily, let’s say we want to execute a similar syntax for column-name-array also. i.e. df.loc[[False, False, False, True, False, True, True, False, False], [False, True, False, True]]. This syntax is again same, we want 3,5,6 row-names and vegetables and legumes as the columns. the [False, True, False, True] indicates that fruits is false, vegetables is true, grains is false and legumes is true. This results in
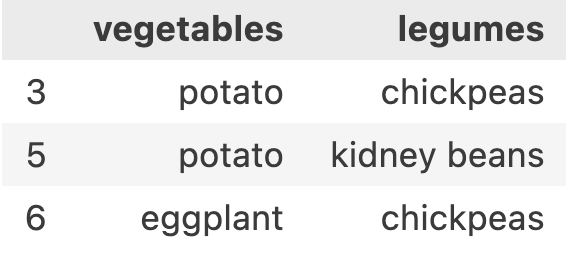
Another interesting thing that you may have noticed is that I can use any way of providing row-name-array and column-name-array. I can use [3,5,6] row-name-array with ['vegetables', 'legumes']. I can use [True, False, ....] with ['vegetables', ...]. I can use [3,5,6] with [True, False, ...] and I can use [True, False, ...] with [True, False, ...].
Fun part is that I can also use row-name with column-name-array and row-name-array with column-name too. For example df.loc[1, [False, True, False, True]] will result in a Series.

Come to think of it, iloc also allows a mix and match of row-number/row-range and column-number/column-range.
Let’s sum it up now.
df[a], in this a can be
- column name => all rows of specific column
- range (python list type slicing syntax) => a bunch of rows
- all-row-boolean-array => specific rows, all columns
iloc[a,b], in this both a and b can be
- a digit (row-number or column-number) => specific row/column
- range (python list type slicing syntax) => a bunch of row/column
- all-row/column-boolean-array (True, False, ..) => specific rows/columns => this array has to be a python list NOT
Series.
loc[a,b] in this both a and b can be
- name (row-name or column-name) => specific row/column
- name-array (row-name-array or column-name-array) => more than one rows/columns
- all-row/column-boolean-array (True, False, ..) => specific rows/columns
What do we mean by [True, True, False, … etc.]?
As promised, we will now talk about this way of passing all-row/column-boolean-array. And what does it mean. To be quick, its filters.
In our data, let’s say we want to filter all the meals which had apple in it.
df['fruits'] == 'apple' this will result in a Series of [True, False, True, False..]
Let’s write it in a better way. I want all rows and columns where apple was a part of the meal
condition = df['fruits'] == 'apple'
df[condition]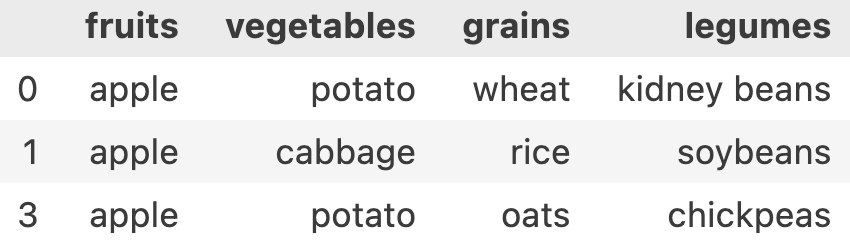
df.iloc[condition.tolist(), :] //since condition is pandas-Series, we need to convert it to python list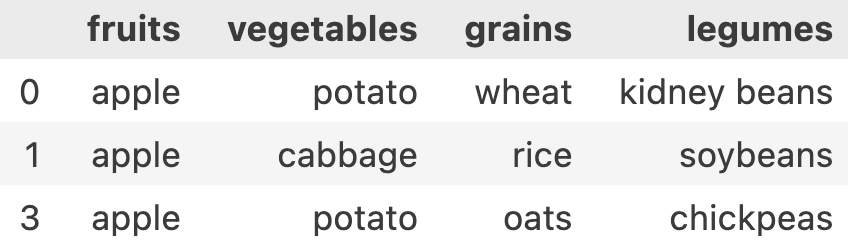
condition = df['fruits'] == 'apple'
df.loc[condition.tolist(), :]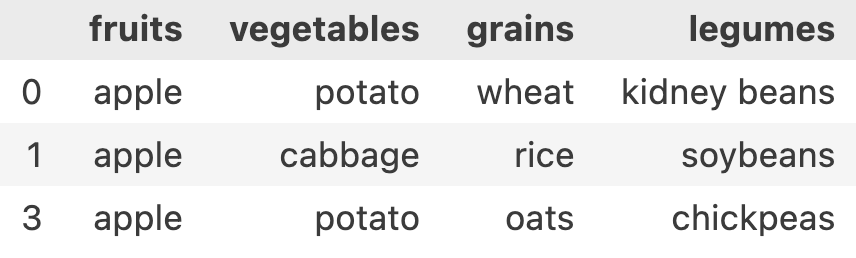
Summary
df[a], in this a can be
- column name => all rows of specific column
- range (python list type slicing syntax) => a bunch of rows
- all-row-boolean-array => specific rows, all columns
iloc[a,b], in this both a and b can be
- a digit (row-number or column-number) => specific row/column
- range (python list type slicing syntax) => a bunch of row/column
- all-row/column-boolean-array (True, False, ..) => specific rows/columns => this array has to be a python list NOT
Series.
loc[a,b] in this both a and b can be
- name (row-name or column-name) => specific row/column
- name-array (row-name-array or column-name-array) => more than one rows/columns
- all-row/column-boolean-array (True, False, ..) => specific rows/columns

Found your learning to be very interesting, had a great satisfaction knowing more about different types of Data frame functions supported by Python while coding.
Had a great learning experience.
Thanks Viraj!
very well simplified as to why we need tabular structure
Thanks Ashutosh!